Data & Analysis Standards
Genomic data can provide an invaluable source of information to understand pathogen evolution, identify patterns of transmission, and characterize phenotypes such as drug resistance and immune escape. For eukaryotic pathogens, larger genomes, sexual recombination, and complicated transmission dynamics including polyclonal infections have historically limited the use of genomic data for many of these applications. However, recent laboratory developments, including multiplexed targeted sequencing, have rapidly increased the pace of genomic data generation for eukaryotic pathogens.
Fundamental differences in the biology and transmission of infections caused by these pathogens render many of the genomic data and analysis tools developed for other organisms (primarily humans, viruses, and bacteria) difficult or impossible to use. As a result, many research efforts have needed to rely on bespoke methods for processing and analysis, limiting the reusability of data, the accuracy and reproducibility of results, and more generally the productivity of scientists studying eukaryotic pathogens. There is a need to develop software and computational tools to process, store, share and analyze these data in a way which sets standards, encourages innovation, and facilitates scientific discovery.
Please provide your feedback, sign up to get involved, or stay informed using our feedback form here!
Proposed standards for targeted amplicon data and metadata
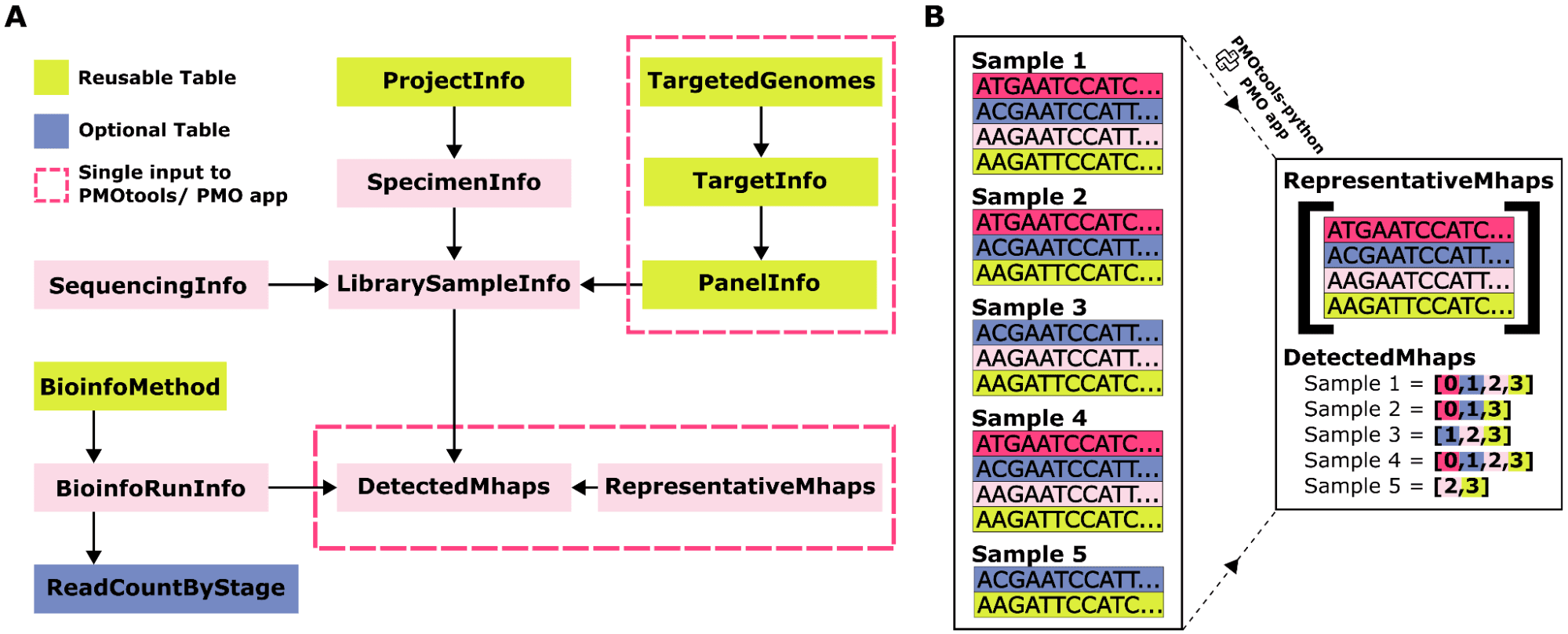
With generation of amplicon sequencing data for Plasmodium accelerating, there is a timely opportunity to create shared resources to disseminate, reuse, and analyze these data. However, there is currently no standard for lossless representation of microhaplotypes derived from these approaches nor for associated laboratory, bioinformatic, and clinical metadata.
A standardized format for microhaplotype data would facilitate data sharing, including development of appropriate repositories, along with transparency and reproducibility of analysis. Standardization at this central step in analysis would also allow for alignment of downstream tools, increasing incentives to develop robust, reusable software and allow cross-study analyses.
We propose the Portable Microhaplotype Object (PMO), a single, relational data structure using JSON as a portable file. This approach allows for a design which is efficient, lightweight, and flexible, organizing metadata together with genetic data.
Work on Data Analysis Standards
To fully utilize the benefits of the proposed standardized data format, we also need a modular software toolkit for analyzing targeted sequencing data. Our proposed work includes:
- Landscaping existing analytical tools, assessing their performance, documentation, and compatibility, and highlighting areas for improvement or gaps that need to be filled.
- Benchmarking tools in collaboration with the broader community to establish standardized evaluation methods.
- Enhancing the reliability, speed, and documentation where needed for priority tools.
- Developing robust end-to-end workflows that integrate tools for common research use cases.
- Hosting these modules and workflows on a scalable, interactive platform.

In December 2023, members of the PlasmoGenEpi community gathered in Baltimore for RADISH23 (Reproducibility, Accessibility, Documentation, and Inter-operability Standards Hackathon 2023). The event aimed to improve the ease of malaria genomic analysis.
Key work included:
- Landscaping 40 tools, with detailed evaluation and tutorials for 17 priority tools.
- Populating the PlasmoGenEpi R-universe for easy installation.
- Launching the PGEforge website to centralize resources.
- Uploading and documenting 4 canonical datasets to use as input to multiple tools.
- Developing the PGEhammer package for data wrangling.
- Strengthening 7 existing packages; MIPanalyzer, PlasmoSim, Pixelate, IsoRelate, hmmIBD, FreqEstimationModel, RAMBLER (now GLAM).
- Establishing suggested software standards and measurable criteria for tool benchmarking.
- Drafting major workflows and a draft paper.
 The website, including the tutorials and information mentioned throughout, can be accessed here!
The website, including the tutorials and information mentioned throughout, can be accessed here! The PMO schema and documentation can be accessed here!
The PMO schema and documentation can be accessed here!